Aptagen, LLC
GBM128 (ID# 7943)

DNA
Glioblastoma
Cells
20 nM (reported value)
The washing buffer (WB) used contained 4.5 g/L glucose, and 5 mM MgCl2 in PBS (pH = 7.4). The binding buffer used during selection was prepared by adding yeast tRNA (0.1 mg/mL) and BSA (1 mg/mL) to washing buffer to reduce background binding.
4°C
NA If the oligo is a known aptamer sequence: For binding studies, perform a refolding protocol to ensure proper function (i.e. binding to antigen or target). Refer to the aptamer reference source for the appropriate refolding parameters and binding conditions. Note: it is unknown whether aptamer functions properly without refolding.
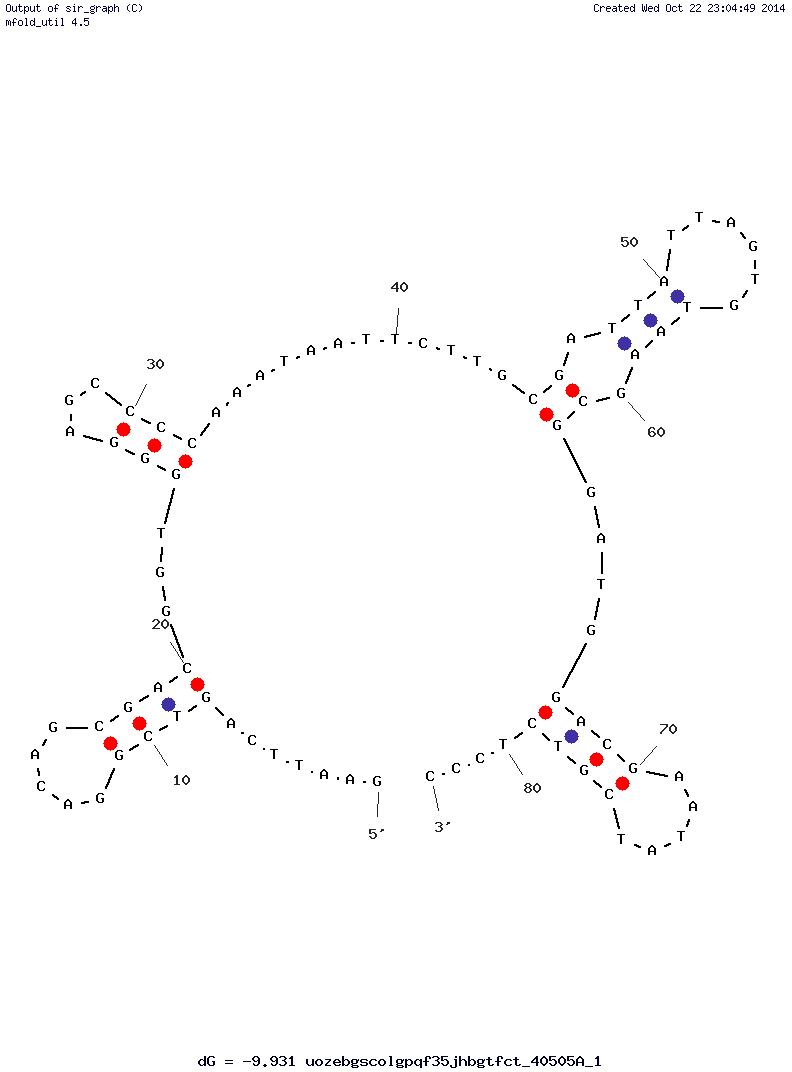
dGpdApdAp dTpdTpdCp dApdGpdTp dCpdGpdGp dApdCpdAp dGpdCpdGp dApdCpdGp dGpdTpdGp dGpdGpdAp dGpdCpdCp dCpdCpdAp dApdApdTp dApdApdTp dTpdCpdTp dTpdGpdCp dGpdApdTp dTpdApdTp dTpdApdGp dTpdGpdTp dApdApdGp dCpdGpdGp dApdTpdGp dGpdApdCp dGpdApdAp dTpdApdTp dCpdGpdTp dCpdTpdCp dCpdCp


83
25689.73
812600
Note: Information on this aptamer oligo was obtained from the literature and hasn't been validated by Aptagen.
Kang D, Wang J, Zhang W, Song Y, Li X, et al. (2012) Selection of DNA Aptamers against Glioblastoma Cells with High Affinity and Specificity. PLoS ONE 7(10): e42731. doi:10.1371/journal.pone.0042731
Have your aptamer oligo synthesized ORDER



We are always looking for ways to improve. Please tell us what you think.