Aptagen, LLC
Apt02 (ID# 9006)

RNA
ITGAV
Protein
359 nM (reported value)
Starting RNA pools (40 mg; 2 nmol) were denatured at 95 °C for 5 min in 100 ml SELEX buffer (145 mM NaCl,
5.4 mM KCl, 0.8 mM MgCl2, 1.8 mM CaCl2, 20 mM Tris-HCl pH 7.6), followed by cooling on ice. The resultant RNAs were further incubated at 37 °C for 10 min, and made up to 3 ml with Cell-SELEXblocking buffer (0.1 mg/ml yeast tRNA, 0.1% bovine serum albumin [BSA], 2 mg/ml D-glucose in SELEX buffer).
37 °C
NA If the oligo is a known aptamer sequence: For binding studies, perform a refolding protocol to ensure proper function (i.e. binding to antigen or target). Refer to the aptamer reference source for the appropriate refolding parameters and binding conditions. Note: it is unknown whether aptamer functions properly without refolding.
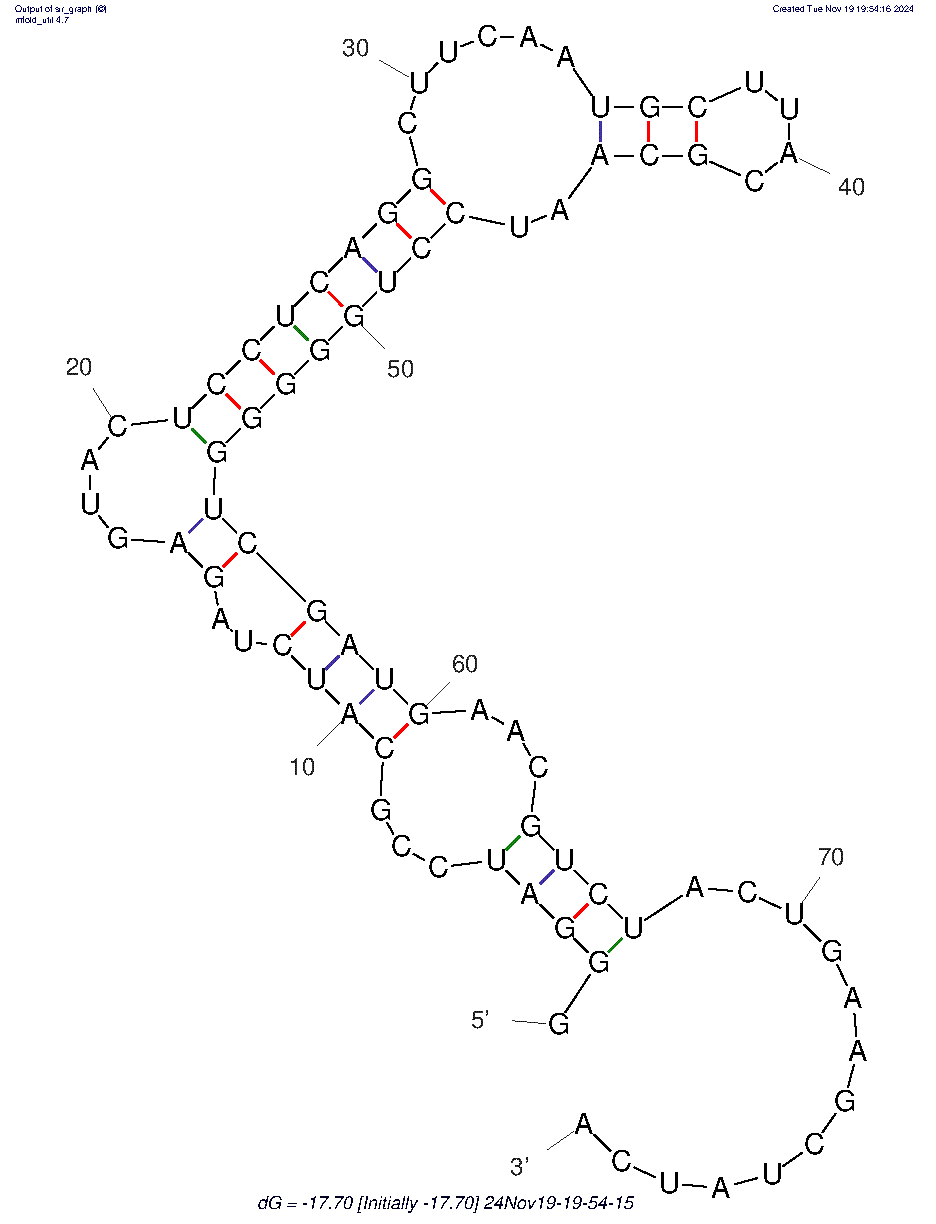
rGprGprGprAprUprCprCprGprCprAprUprCprUprAprGprAprGprUprAprCprUprCprCprUprCprAprGprGprCprUprUprCprAprAprUprGprCprUprUprAprCprGprCprAprAprUprCprCprUprGprGprGprGprGprUprCprGprAprUprGprAprAprCprGprUprCprUprAprCprUprGprAprAprGprCprUprAprUprCprAp


80
24349.4 g/mole
736300 L/(mole·cm)
Note: Information on this aptamer oligo was obtained from the literature and hasn't been validated by Aptagen.
Takahashi M, Sakota E, Nakamura Y. The efficient cell-SELEX strategy, Icell-SELEX, using isogenic cell lines for selection and counter-selection to generate RNA aptamers to cell surface proteins. Biochimie. 2016 Dec;131:77-84. doi: 10.1016/j.biochi.2016.09.018. Epub 2016 Oct 1. PMID: 27693080.
Have your aptamer oligo synthesized ORDER



We are always looking for ways to improve. Please tell us what you think.