Aptagen, LLC
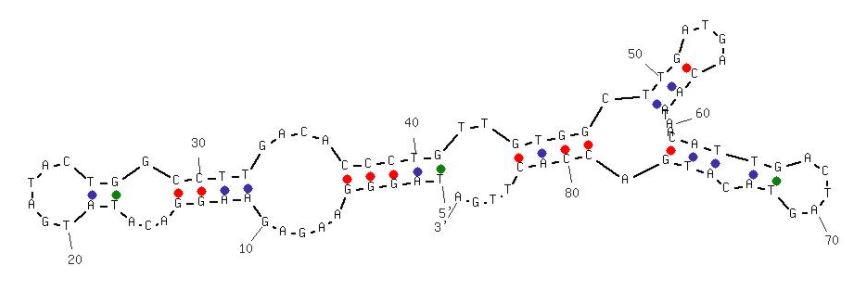
EF508 (ID# 8096)

DNA
Enterococcus faecalis bacteria
Other
37 nM (reported value)
10 mM of Tris–EDTA bufer (pH 8.5)
22°C
NA If the oligo is a known aptamer sequence: For binding studies, perform a refolding protocol to ensure proper function (i.e. binding to antigen or target). Refer to the aptamer reference source for the appropriate refolding parameters and binding conditions. Note: it is unknown whether aptamer functions properly without refolding.
5’-dTpdApdGpdGpdGpdApdApdGpdApdGpdApdApdGpdGpdApdCpdApdTpdApdTpdGpdApdTpdApdCpdTpdGpdGpdCpdCpdTpdTpdGpdApdCpdApdCpdCpdCpdTpdGpdTpdTpdGpdTpdGpdGpdCpdTpdTpdGpdApdTpdGpdApdCpdApdApdTpdApdApdCpdApdTpdTpdGpdApdCpdTpdApdGpdTpdApdCpdApdTpdGpdApdCpdCpdApdCpdTpdTpdGpdAp-3’


86
26627.3
853800
Note: Information on this aptamer oligo was obtained from the literature and hasn't been validated by Aptagen.
Kolm C, Cervenka I, Aschl UJ, et al. DNA aptamers against bacterial cells can be efficiently selected by a SELEX process using state-of-the art qPCR and ultra-deep sequencing. Sci Rep. 2020;10(1):20917. Published 2020 Dec 1. doi:10.1038/s41598-020-77221-9
Have your aptamer oligo synthesized ORDER



We are always looking for ways to improve. Please tell us what you think.